#| '!! shinylive warning !!': |
#| shinylive does not work in self-contained HTML documents.
#| Please set `embed-resources: false` in your metadata.
#| standalone: true
#| components: [viewer]
#| viewerHeight: 750
from shiny import App, Inputs, Outputs, Session, render, ui
from shiny import reactive
import numpy as np
from pathlib import Path
import matplotlib.pyplot as plt
from scipy.integrate import odeint
app_ui = ui.page_fluid(
ui.layout_sidebar(
ui.sidebar(
ui.input_slider(id="R_0",label="R_0",min=0.0,max=5.0,value=1.2,step=0.001),
ui.input_slider(id="a",label="a",min=0.0,max=5.0,value=1.2,step=0.001),
ui.input_slider(id="C",label="C",min=0.0,max=5.0,value=1.2,step=0.001),
ui.input_slider(id="N_0",label="N_0 (Initial condition)",min=0.0,max=4.0,value=0.5,step=0.01),
ui.input_slider(id="P_0",label="P_0 (Initial condition)",min=0.0,max=4.0,value=0.5,step=0.01),
ui.input_slider(id="T",label="Number of iterations",min=0.0,max=60.0,value=20.0,step=1.0),
),
ui.output_plot("plot"),
),
)
def server(input, output, session):
@render.plot
def plot():
fig, ax = plt.subplots()
#ax.set_ylim([-2, 2])
# Filter fata
R_0=float(input.R_0())
a=float(input.a())
C=float(input.C())
T=int(input.T())
N_0=float(input.N_0())
P_0=float(input.P_0())
par=[R_0,a,C]
init_cond=[N_0,P_0]
def nicholsonbaileyrhs(x,par):
f=np.zeros_like(x,dtype=float)
N=x[0]
P=x[1]
R_0=par[0]
a=par[1]
C=par[2]
f_nb=np.exp(-a*P)
f[0]=R_0*N*f_nb
f[1]=C*N*(1-f_nb)
return f
# Define rhs of logistic map
# this function computes the solutio to the difference equation. It also stores solution in format for cobweb plotting
def SolveSingleDiffAndCobweb(t,rhs,init_cond,par):
# Initialise solution vector
N = np.zeros_like(t,dtype=float)
P = np.zeros_like(t,dtype=float)
N[0]=init_cond[0]
P[0]=init_cond[1]
# loop over time and compute solution.
for i in t:
if i>0:
sol=[N[i-1],P[i-1]]
rhs_eval=rhs(sol,par)
N[i]=rhs_eval[0]
P[i]=rhs_eval[1]
return N, P
# Define discretised t domain
t = np.arange(0, T, 1)
# define initial conditions
# Compute numerical solution of ODEs
N,P = SolveSingleDiffAndCobweb(t,nicholsonbaileyrhs,init_cond,par)
# Plot results
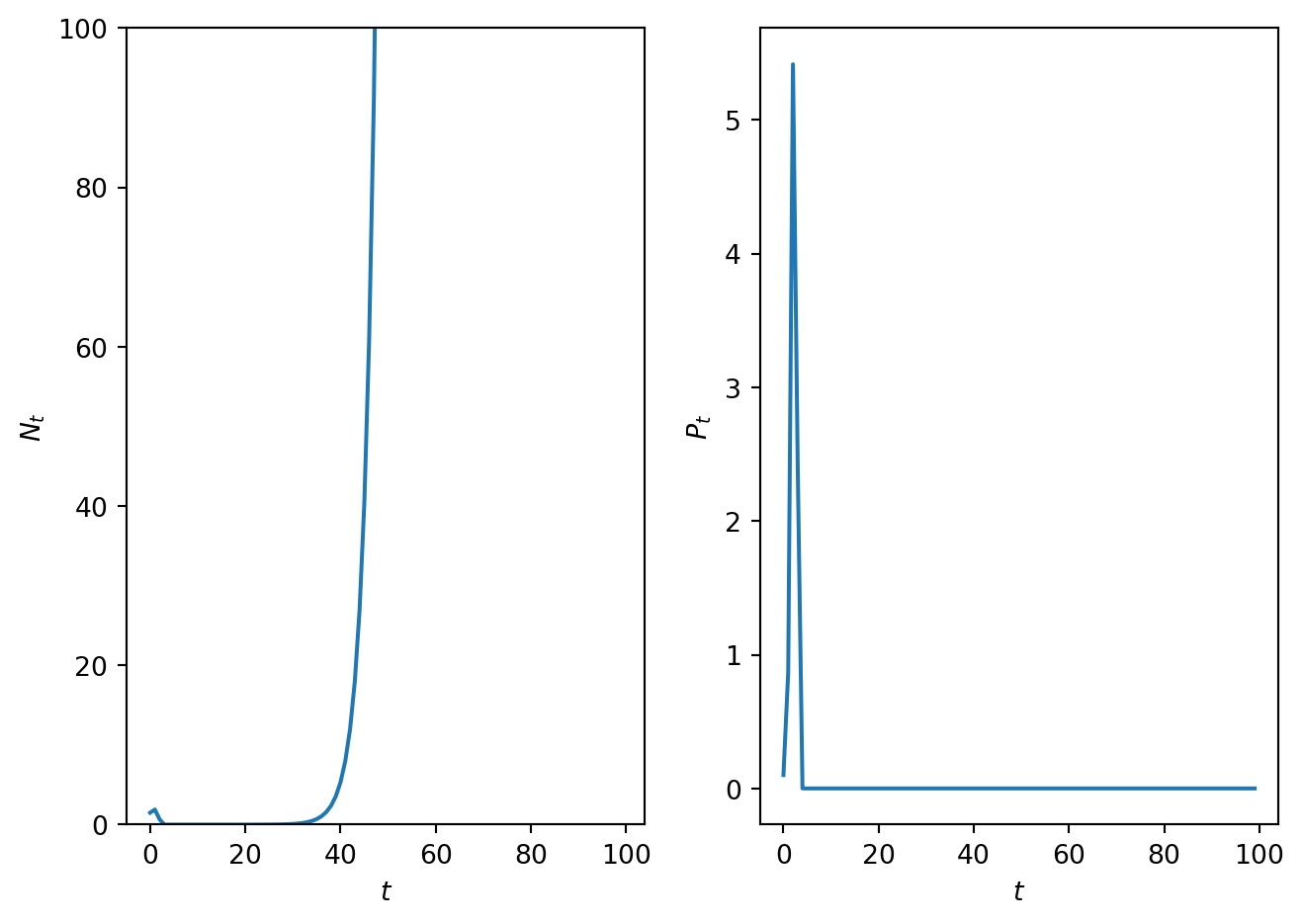
ax.plot(t,N,t,P)
ax.set_xlabel('$t$')
ax.legend(['$N_t$','$P_t$'])
#ax.set_ylim([0,max_sol])
ax.set_title('Time series')
plt.grid()
plt.show()
app = App(app_ui, server)